
tl;dr
You can use R to extract running details from a downloaded of your Apple Health data. My old code broke, so I re-wrote it.
On your marks
In 2021 I extracted my running activities from my Apple Health data using the {xml2} package. You can read there for some theory and background.
At that point I’d been running for one year. I’m nearly at 500 runs1, so I thought I would re-execute my code with the latest data. Alas, the original code no longer works because Apple seems to have updated the format of the XML file they provide.
So I’ve written a new function that takes a path to the zipped download of my Apple Health data and outputs a dataframe of time and distance data, with one row per run.
Get set
I followed the same steps as before to get my Apple Health data off my phone.
I smashed together a quick function to unzip the file to a temporary location and then extract workout data using the the {xml2} package. There’s a bit of base R wrangling to output a dataframe with a row per run workout, focusing on total time and distance.
Click to expand the function definition
get_run_distances <- function(zip_path) {
# Unzip Apple Health export to temporary location
message("Unzipping and reading XML")
temp <- tempdir()
unzip(zipfile = zip_path, exdir = temp)
xml_in <- xml2::read_xml(file.path(temp, "apple_health_export", "export.xml"))
unlink(temp)
# Isolate workouts only and convert to an R list object
message("Isolating workouts from XML")
wo_in <- xml2::xml_find_all(xml_in, "//Workout") |> xml2::as_list()
# Pre-allocate a list to be filled with output data
wo_total <- length(wo_in)
wo_out <- vector("list", wo_total)
# For each viable workout, extract the details
message("Iterating over workouts to extract run data")
for (wo_n in seq(wo_total)) {
# Extract details for current workout
wo <- wo_in[[wo_n]]
wo_attrs <- attributes(wo) # the data is stored as attributes
is_run <-
wo_attrs[["workoutActivityType"]] == "HKWorkoutActivityTypeRunning"
# If the workout wasn't a run, then skip to the next workout
if (!is_run) next
# if it is a run, then extract the data to a single-row dataframe
if (is_run) {
# There can be more than one element named 'WorkoutStatistics'. We want to
# get the one with distance information and extract the details.
wo_stats <- wo[grep("WorkoutStatistics", names(wo))]
wo_stats_types <- lapply(wo_stats, \(x) attr(x, c("type")))
dist_type <- "HKQuantityTypeIdentifierDistanceWalkingRunning"
dist_index <- which(wo_stats_types == dist_type)
wo_dist <- wo_stats[[dist_index]]
# Prepare single-row dataframe and add to the pre-allocated list
wo_details <- data.frame(
source = wo_attrs[["sourceName"]],
start = as.POSIXct(wo_attrs[["startDate"]]),
end = as.POSIXct(wo_attrs[["endDate"]]),
distance_km = attr(wo_dist, "sum") |> as.numeric() |> round(2)
)
wo_details[["duration_s"]] <-
as.numeric(wo_details[["end"]] - wo_details[["start"]], units = "secs")
wo_out[[wo_n]] <- wo_details
}
}
# Convert to dataframe, select columns
message("Combining data")
wo_out_df <- do.call(rbind, wo_out)
wo_out_df[, c("source", "start", "end", "duration_s", "distance_km")]
}I won’t go through it line by line, but there’s some commentary to explain what’s happening at each step. It does what I need it to do for now, but no doubt there’s some refactoring to be done.
There’s a few things to note:
- I’m more comfortable handling R objects, so I converted early to a list with
xml2::as_list(). Awkwardly, the data in the list object was stored as attributes to each element. - The distance data is stored in an element called ‘WorkoutStatistics’, but more than one element will have this name. We first have to isolate the element that is of the correct type, which has the name ‘HKQuantityTypeIdentifierDistanceWalkingRunning’.
- I converted the start and end variables to datetime class (POSIXct) and subtracted one from the other to get the duration of the run. This yields the ‘difftime’ class that can be converted to seconds with
as.numeric()and the argumentunits = "secs". - There’s no input handling, because this was quick and for ‘fun’, lol.
Go
So, to use the function you pass a path to where your zipped Apple Health export lives. Mine is in my ‘Downloads’ folder.
runs <- get_run_distances("~/Downloads/export.zip")## Unzipping and reading XML## Isolating workouts from XML## Iterating over workouts to extract run data## Combining dataI recorded all my runs with the Nike Run Club app, so I’ll filter out duplicates where I dual-recorded with Apple’s Workout app. I think I accidentally started the app by mistake a couple of times, so we’ll only grab runs of over 1 km. I’ll also convert the seconds to a friendlier-looking ‘period’ class using {lubridate}2.
Here’s the most recent few:
runs <- runs[runs$source == "Nike Run Club" & runs$distance_km > 1, ]
runs$duration <- lubridate::seconds_to_period(runs$duration_s)
runs <- runs[, c("start", "distance_km", "duration")]
row.names(runs) <- NULL
tail(runs)## start distance_km duration
## 490 2023-05-27 10:57:37 6.91 32M 22S
## 491 2023-05-28 10:21:40 10.15 48M 59S
## 492 2023-05-31 17:10:27 5.26 23M 23S
## 493 2023-06-02 16:51:47 6.57 30M 36S
## 494 2023-06-04 08:35:03 10.16 47M 59S
## 495 2023-06-07 16:42:26 12.67 1H 0M 16SFor my own tracking purposes, I’ve run:
- 495 times
- for a total distance of 4053 km
- for a total duration of about 14 days
And I can recreate one of the plots from the old post while we’re here:
plot(
x = runs$start,
y = runs$distance_km,
las = 1, # rotate y-axis labels
main = "Runs captured with Nike Run Club in Apple Health",
xlab = "Date",
ylab = "Distance (km)"
)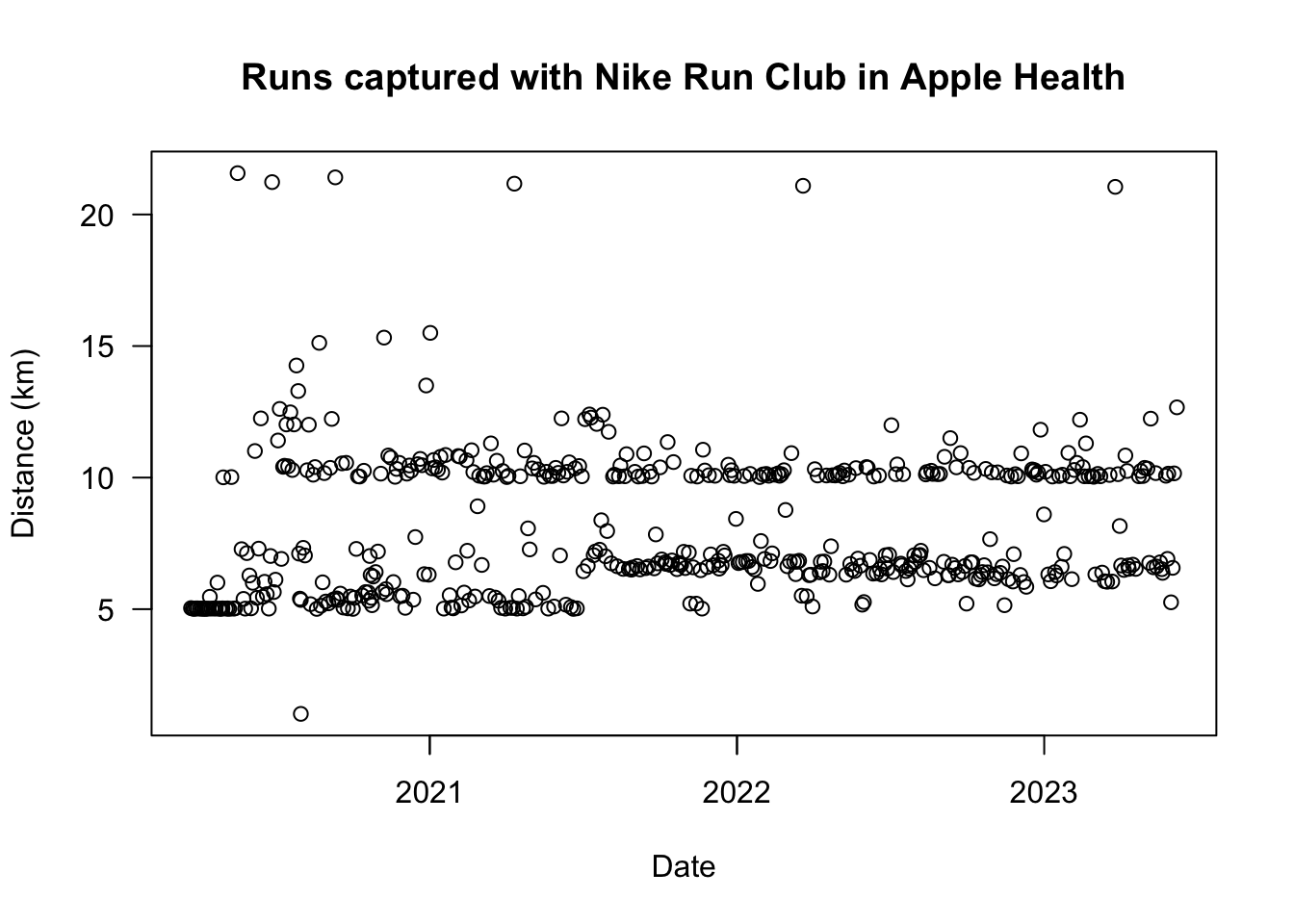
Some patterns are obvious. For example, there’s lots of 5 km runs until about mid-2021, when it hops to more like 7 km. That’s when I started running for 30 mins at a time, rather than for 5 km specifically.
I’m pretty happy at these two distances, obviously, but maybe I should do more 21.1 km half-marathons. Or a full marathon? No no, that’s foolish: it would expand my y-axis too much and make it harder to observe patterns at shorter distances, amirite.
Session info
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.2.0 (2022-04-22)
## os macOS Big Sur/Monterey 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Europe/London
## date 2023-06-11
## pandoc 2.19.2 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## blogdown 1.9 2022-03-28 [1] CRAN (R 4.2.0)
## bookdown 0.26 2022-04-15 [1] CRAN (R 4.2.0)
## bslib 0.3.1 2021-10-06 [1] CRAN (R 4.2.0)
## cli 3.6.1 2023-03-23 [1] CRAN (R 4.2.0)
## digest 0.6.31 2022-12-11 [1] CRAN (R 4.2.0)
## evaluate 0.20 2023-01-17 [1] CRAN (R 4.2.0)
## fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.2.0)
## generics 0.1.3 2022-07-05 [1] CRAN (R 4.2.0)
## highr 0.10 2022-12-22 [1] CRAN (R 4.2.0)
## htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.2.0)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.2.0)
## jsonlite 1.8.4 2022-12-06 [1] CRAN (R 4.2.0)
## knitr 1.42 2023-01-25 [1] CRAN (R 4.2.0)
## lubridate * 1.9.2 2023-02-10 [1] CRAN (R 4.2.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.2.0)
## rlang 1.1.1 2023-04-28 [1] CRAN (R 4.2.0)
## rmarkdown 2.14 2022-04-25 [1] CRAN (R 4.2.0)
## rstudioapi 0.14 2022-08-22 [1] CRAN (R 4.2.0)
## sass 0.4.1 2022-03-23 [1] CRAN (R 4.2.0)
## sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.2.0)
## timechange 0.1.1 2022-11-04 [1] CRAN (R 4.2.0)
## xfun 0.37 2023-01-31 [1] CRAN (R 4.2.0)
## yaml 2.3.7 2023-01-23 [1] CRAN (R 4.2.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.2/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────Yeah, I could’ve waited to post this after I’d actually reached 500 runs, but I like arbitrary chaos.↩︎
{lubridate} is handy for time handling for many reasons. Here it’s helpful because it can resolve minutes and seconds (e.g.
21M 30S) instead of the decimal minutes (e.g.21.5) in a difftime object.↩︎